Forecasting Google Scholar statistics for researchers at the Centre for Human Brain Health
Note: this guide is also available on the corresponding GitHub repository, where all the required code is freely available.
This is a detailed guide for performing the extraction and forecasting of Google Scholar data for researchers at the Centre for Human Brain Health (CHBH), University of Birmingham.
Specficially, this involves:
- Scraping a list of researchers at the CHBH from the CHBH website (using
beautifulsoup4). - Extracting Google Scholar statistics including citation data (using
serpapi). - Forecasting citations for the years 2024, 2025 and 2026 (using
prophet).
Extracting data from the CHBH website
The first step is to get a list of names for researchers at the CHBH. We could do this in a bunch of different ways, but one way is to use beautifulsoup4 to extract names from six pages on the CHBH website that list senior researchers.
Here is how the webpages display the researcher’s information (six pages in total):
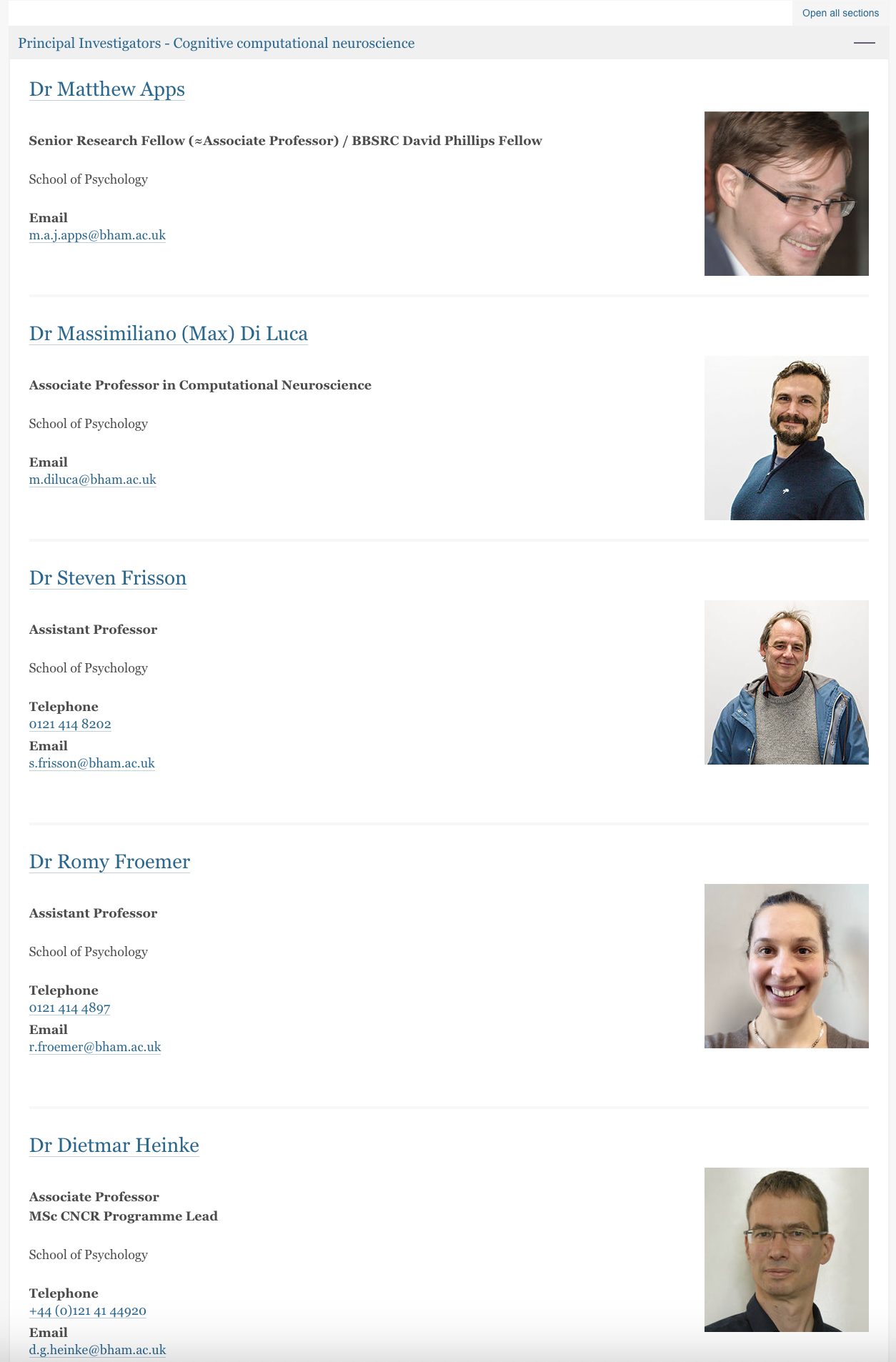
And here is the code which extract the names scrape_chbh.py which it iterates over the list of URLs, and extracts names using the BeautifulSoup library to parse the HTML content. Importantly, the script also filters and cleans these names by removing titles like “Dr” and “Professor”.
import requests
from bs4 import BeautifulSoup
import json
def scrape_names_from_page(url):
response = requests.get(url)
soup = BeautifulSoup(response.content, 'html.parser')
# Find all elements that contain names - this will need to be customized based on the page's HTML structure
name_elements = soup.find_all('h2') # Assuming names are within <h2> tags, change as needed
names = [elem.get_text(strip=True) for elem in name_elements]
return names
# List of URLs to scrape
urls = [
"https://www.birmingham.ac.uk/research/centre-for-human-brain-health/chbh-research-themes/lifespan-and-brain-health",
"https://www.birmingham.ac.uk/research/centre-for-human-brain-health/chbh-research-themes/cognitive-computational-neuroscience",
"https://www.birmingham.ac.uk/research/centre-for-human-brain-health/chbh-research-themes/social-interaction-and-communication",
"https://www.birmingham.ac.uk/research/centre-for-human-brain-health/chbh-research-themes/learning-memory-and-performance",
"https://www.birmingham.ac.uk/research/centre-for-human-brain-health/chbh-research-themes/awareness-consciousness-and-sleep",
"https://www.birmingham.ac.uk/research/centre-for-human-brain-health/chbh-research-themes/neuroimaging-methods-and-ai"
]
# Initialize an empty set to hold all unique names across themes
unique_names = set()
# Scrape names from each URL
for url in urls:
names = scrape_names_from_page(url)
for name in names:
if 'Dr ' in name or 'Professor ' in name:
# Clean name and add to the set of unique names
clean_name = name.replace('Dr ', '').replace('Professor ', '').strip()
unique_names.add(clean_name)
This gives us a list of CHBH researchers (chbh_names.json).
{
"chbh-investigators": [
"Alan Wing",
"Andrea Krott",
"Kim Shapiro",
"Craig McAllister",
"Clare Anderson",
"Rachel Upthegrove",
"Dietmar Heinke",
"Lei Zhang",
"Magda Chechlacz",
"Joseph Galea",
"Massimiliano (Max) Di Luca",
"Jennifer Cook",
"Matthew Apps",
"KyungMin An",
"Katja Kornysheva",
"Andrew J. Bremner",
"Ali Mazaheri",
"Hamid Dehghani",
"Damian Cruse",
"Patricia Lockwood",
"Ned Jenkinson",
"Tom Rhys Marshall",
"Chris Miall",
"Stephane De Brito",
"Romy Froemer",
"Anna Kowalczyk",
"Suzanne Higgs",
"Sarah Aldred",
"Andrew Bagshaw",
"Rickson C. Mesquita",
"Martin Wilson",
"Davinia Fernández-Espejo",
"Andrew Quinn",
"Hyojin Park",
"Karen Mullinger",
"Arkady Konovalov",
"Felipe Orihuela-Espina",
"Carmel Mevorach",
"Paul Muhle-Karbe",
"Clayton Hickey",
"Katrien Segaert",
"Nick Holmes",
"Sam Lucas",
"Ole Jensen",
"Barbara Pomiechowska",
"Jian Liu",
"Steven Frisson"
]
}
Extracting Google Scholar data using SerpAPI
SerpAPI is a tool that provides developers with an API to scrape and parse search engine results pages (SERPs) from various search engines like Google, Bing, and others, automating the process of extracting search data.
SerpAPI offers a range of APIs for Google services, including: Google Search API, Google Images API, Google News API, Google Shopping API, and relevant for this exercise, Google Scholar API. It has a free tier, but restricts users to only 100 calls per month, so use them wisely!
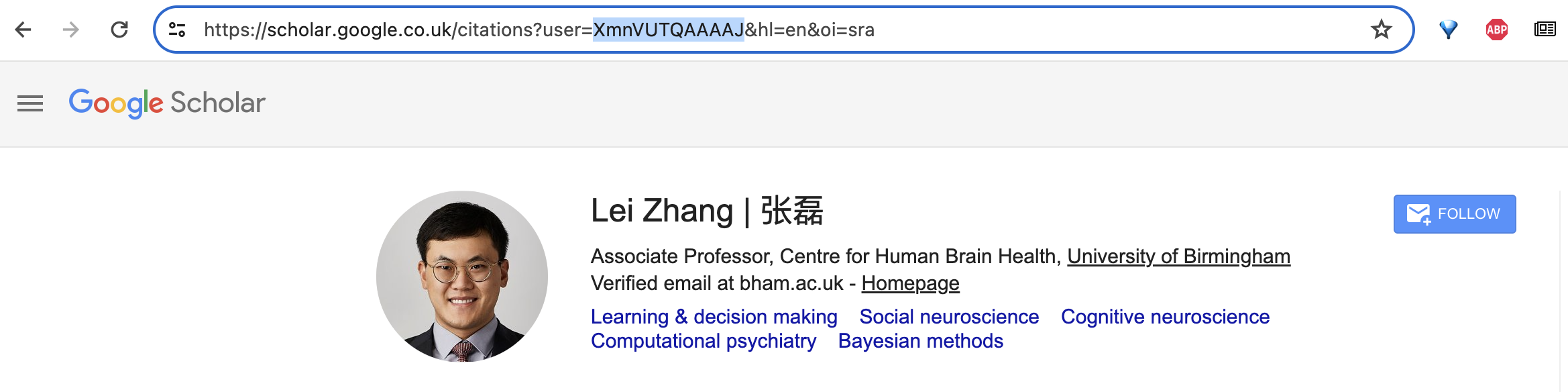
Importantly, SerpAPI works using Google Scholar ID’s and not on the researchers names. You can see the ID within the URL when on the researchers profile page. For example, Lei Zhang’s ID is XmnVUTQAAAAJ.
I could have written a script to extract this for the researchers using SerpAPI, but given that there are 42 of them, and that I am limited to 100 requests per month (and I’ll need at least 42 for the actual extraction), I just decided to do this manually, creating a csv file names_and_citation_ids.csv which I then filtered by removing those with no Scholar ID. The final data is filtered_names_and_citations_ids.csv which looks like this:
name,citation_id
Alan Wing,JEihq_0AAAAJ
Andrea Krott,Hfg7LVkAAAAJ
Kim Shapiro,RO_OKQwAAAAJ
Craig McAllister,KFT9l3wAAAAJ
Clare Anderson,MHtyzVoAAAAJ
Rachel Upthegrove,Z1kBsNIAAAAJ
Dietmar Heinke,uExP9ScAAAAJ
Lei Zhang,XmnVUTQAAAAJ
Magda Chechlacz,DSIp5Mh1f3oC
Joseph Galea,xVLhU5kAAAAJ
Extracting the Citation statistics
We can now perform the extraction! Here is the important part of the code which does this (extract_scholar_info.py). You will need to provide your own SerpAPI Key for the script to work! The input for this script comes from the CSV file just created named filtered_names_and_citation_ids.csv, and loops through each row, performing the extraction on each citation_id. The script processes these IDs, retrieves the relevant data for each author, and saves this information in a separate JSON file named after the author’s citation ID.
import serpapi
import json
import pandas as pd
def author_results(author_id):
print(f"Extracting author results for author ID: {author_id}")
params = {
"api_key": "SERP_API_KEY", # Replace with your actual SerpApi API key
"engine": "google_scholar_author", # Author results search engine
"author_id": author_id, # Author ID as function input
"hl": "en"
}
search = serpapi.search(params)
results = search # Directly use the 'results'
# Extract the necessary information
thumbnail = results.get("author", {}).get("thumbnail")
name = results.get("author", {}).get("name")
affiliations = results.get("author", {}).get("affiliations")
email = results.get("author", {}).get("email")
website = results.get("author", {}).get("website")
interests = results.get("author", {}).get("interests")
cited_by_table = results.get("cited_by", {}).get("table")
cited_by_graph = results.get("cited_by", {}).get("graph")
public_access_link = results.get("public_access", {}).get("link")
available_public_access = results.get("public_access", {}).get("available")
not_available_public_access = results.get("public_access", {}).get("not_available")
co_authors = results.get("co_authors")
author_results_data = {
"thumbnail": thumbnail,
"name": name,
"affiliations": affiliations,
"email": email,
"website": website,
"interests": interests,
"cited_by_table": cited_by_table,
"cited_by_graph": cited_by_graph,
"public_access_link": public_access_link,
"available_public_access": available_public_access,
"not_available_public_access": not_available_public_access,
"co_authors": co_authors
}
# Save the data to a JSON file named after the author_id
with open(f"{author_id}.json", 'w', encoding='utf-8') as f:
json.dump(author_results_data, f, ensure_ascii=False, indent=4)
print(f"Data saved to {author_id}.json")
return author_results_data
The individual JSON files contains information contained within one’s Scholar Page. We can do a whole range of potential analyses, but we are interested in just using the citation statistics for now.
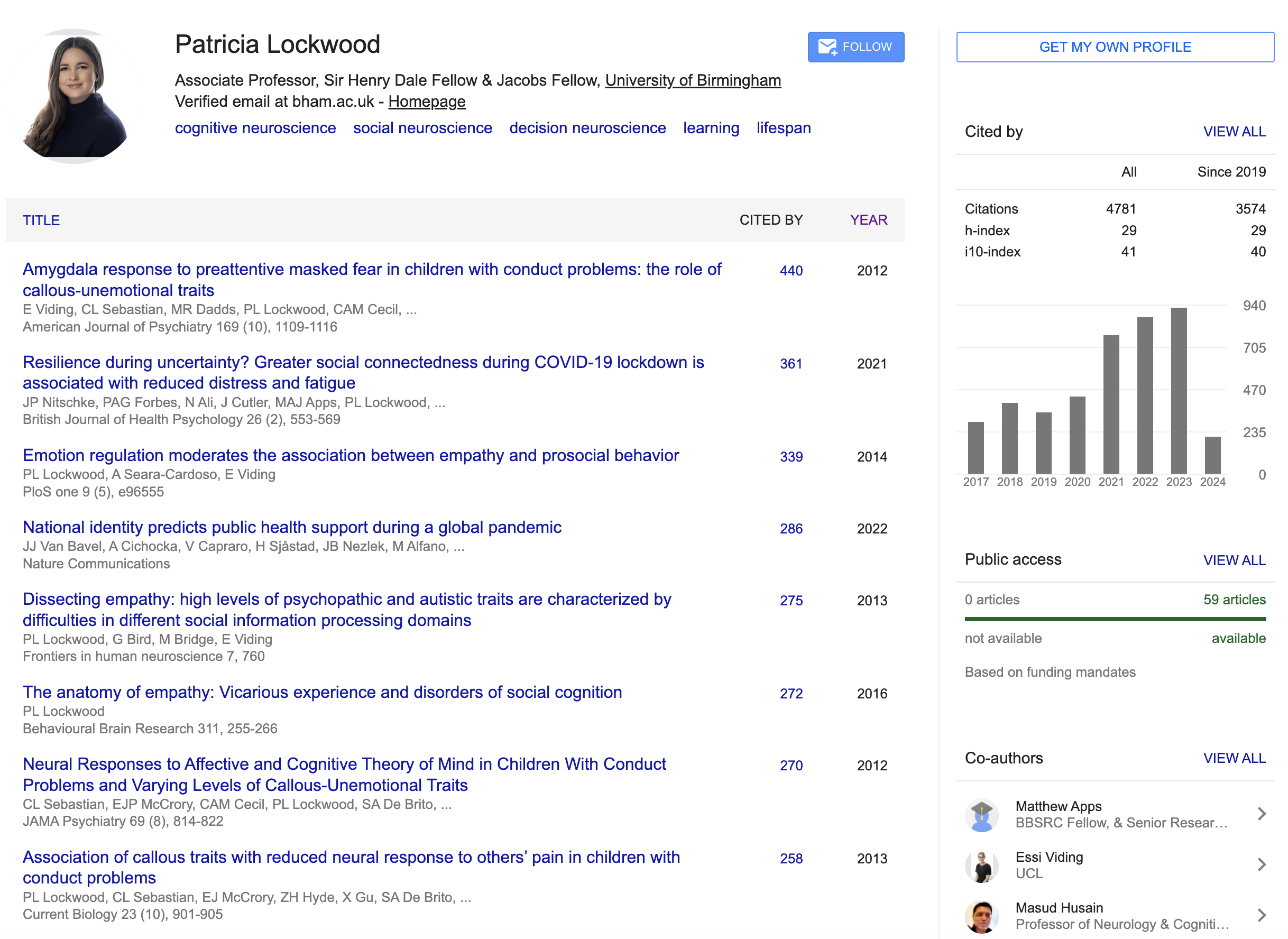
{
"thumbnail": "https://scholar.googleusercontent.com/citations?view_op=view_photo&user=ynN6pG0AAAAJ&citpid=9",
"name": "Patricia Lockwood",
"affiliations": "Associate Professor, Sir Henry Dale Fellow & Jacobs Fellow, University of Birmingham",
"email": "Verified email at bham.ac.uk",
"website": "http://www.sdn-lab.org/",
"interests": [
{
"title": "cognitive neuroscience",
"link": "https://scholar.google.com/citations?view_op=search_authors&hl=en&mauthors=label:cognitive_neuroscience",
"serpapi_link": "https://serpapi.com/search.json?engine=google_scholar_profiles&hl=en&mauthors=label%3Acognitive_neuroscience"
},
{
"title": "social neuroscience",
"link": "https://scholar.google.com/citations?view_op=search_authors&hl=en&mauthors=label:social_neuroscience",
"serpapi_link": "https://serpapi.com/search.json?engine=google_scholar_profiles&hl=en&mauthors=label%3Asocial_neuroscience"
},
{
"title": "decision neuroscience",
"link": "https://scholar.google.com/citations?view_op=search_authors&hl=en&mauthors=label:decision_neuroscience",
"serpapi_link": "https://serpapi.com/search.json?engine=google_scholar_profiles&hl=en&mauthors=label%3Adecision_neuroscience"
},
{
"title": "learning",
"link": "https://scholar.google.com/citations?view_op=search_authors&hl=en&mauthors=label:learning",
"serpapi_link": "https://serpapi.com/search.json?engine=google_scholar_profiles&hl=en&mauthors=label%3Alearning"
},
{
"title": "lifespan",
"link": "https://scholar.google.com/citations?view_op=search_authors&hl=en&mauthors=label:lifespan",
"serpapi_link": "https://serpapi.com/search.json?engine=google_scholar_profiles&hl=en&mauthors=label%3Alifespan"
}
],
"cited_by_table": [
{
"citations": {
"all": 4773,
"since_2019": 3566
}
},
{
"h_index": {
"all": 29,
"since_2019": 29
}
},
{
"i10_index": {
"all": 41,
"since_2019": 40
}
}
],
"cited_by_graph": [
{
"year": 2013,
"citations": 40
},
{
"year": 2014,
"citations": 103
},
{
"year": 2015,
"citations": 126
},
{
"year": 2016,
"citations": 214
},
{
"year": 2017,
"citations": 289
},
{
"year": 2018,
"citations": 396
},
{
"year": 2019,
"citations": 342
},
{
"year": 2020,
"citations": 431
},
{
"year": 2021,
"citations": 776
},
{
"year": 2022,
"citations": 874
},
{
"year": 2023,
"citations": 924
},
{
"year": 2024,
"citations": 200
}
],
"public_access_link": "https://scholar.google.com/citations?view_op=list_mandates&hl=en&user=ynN6pG0AAAAJ",
"available_public_access": 59,
"not_available_public_access": 0,
"co_authors": [
{
"name": "Matthew Apps",
"link": "https://scholar.google.com/citations?user=ZTqTFbwAAAAJ&hl=en",
"serpapi_link": "https://serpapi.com/search.json?author_id=ZTqTFbwAAAAJ&engine=google_scholar_author&hl=en",
"author_id": "ZTqTFbwAAAAJ",
"affiliations": "BBSRC Fellow, & Senior Research Fellow (=Associate Professor)",
"email": "Verified email at bham.ac.uk",
"thumbnail": "https://scholar.google.com/citations/images/avatar_scholar_56.png"
},
{
"name": "Essi Viding",
"link": "https://scholar.google.com/citations?user=Ydwv2DYAAAAJ&hl=en",
"serpapi_link": "https://serpapi.com/search.json?author_id=Ydwv2DYAAAAJ&engine=google_scholar_author&hl=en",
"author_id": "Ydwv2DYAAAAJ",
"affiliations": "UCL",
"email": "Verified email at ucl.ac.uk",
"thumbnail": "https://scholar.googleusercontent.com/citations?view_op=small_photo&user=Ydwv2DYAAAAJ&citpid=13"
},
{
"name": "Masud Husain",
"link": "https://scholar.google.com/citations?user=_GXoXKMAAAAJ&hl=en",
"serpapi_link": "https://serpapi.com/search.json?author_id=_GXoXKMAAAAJ&engine=google_scholar_author&hl=en",
"author_id": "_GXoXKMAAAAJ",
"affiliations": "Professor of Neurology & Cognitive Neuroscience, Oxford. Editor-in-Chief, Brain",
"email": "Verified email at ndcn.ox.ac.uk",
"thumbnail": "https://scholar.googleusercontent.com/citations?view_op=small_photo&user=_GXoXKMAAAAJ&citpid=34"
},
(and so on)
]
}
We can then simply extract this data across all JSON files by running processing_citation_data.py
import os
import json
import pandas as pd
# List all JSON files in the current directory
json_files = [f for f in os.listdir('.') if os.path.isfile(f) and f.endswith('.json')]
# Initialize a list to store data for each author
authors_data = []
# Process each JSON file
for filename in json_files:
with open(filename, 'r', encoding='utf-8') as file:
data = json.load(file)
# Extracting required information
name = data['name']
citations_all = next((item for item in data['cited_by_table'] if 'citations' in item), {}).get('citations', {}).get('all', 0)
h_index_all = next((item for item in data['cited_by_table'] if 'h_index' in item), {}).get('h_index', {}).get('all', 0)
i10_index_all = next((item for item in data['cited_by_table'] if 'i10_index' in item), {}).get('i10_index', {}).get('all', 0)
# Initialize a dict for the author's citation years data
citation_years = {'name': name, 'citations': citations_all, 'h-index': h_index_all, 'i10-index': i10_index_all}
# Add citation count per year to the dict
for item in data['cited_by_graph']:
year = item['year']
citations = item['citations']
citation_years[f'citations_{year}'] = citations
# Append the author's data to the list
authors_data.append(citation_years)
# Convert the list of data into a DataFrame
df = pd.DataFrame(authors_data)
# Fill NaN values with 0 to indicate years without citations
df.fillna(0, inplace=True)
# Save the DataFrame to a CSV file
df.to_csv('../data/citations_statistics/citation_statistics.csv', index=False, encoding='utf-8')
Which creates the citation_statistics.csv, containing all the citation statistics for all researchers!
Forecasting future citations using Prophet
There are many different approaches we can use to forecast the number of citations a researcher will get in the future. I will be using prophet, a open-source forecasting tool developed by Facebook and designed for ease of use and handling the complexities of daily, weekly, and yearly seasonal patterns.
Note, that I am not using Prophet because it’s the most accurate or even the most suitable for this data, I just wanted to try it out!
Working with the Jupyter Notebook for plotting
The code for subsetting the data, forecasting and plotting the forecasted plots are available in plotting_chbh_statistics.ipynb.
You will need to install a kernel for Jupyter Notebooks within the virtual environment:
python -m ipykernel install --user --name=.venv
and then can run the Jupyter Notebook.
python -m notebook
The notebook firstly plots the csv data as is, generating citation numbers over time for all researchers:
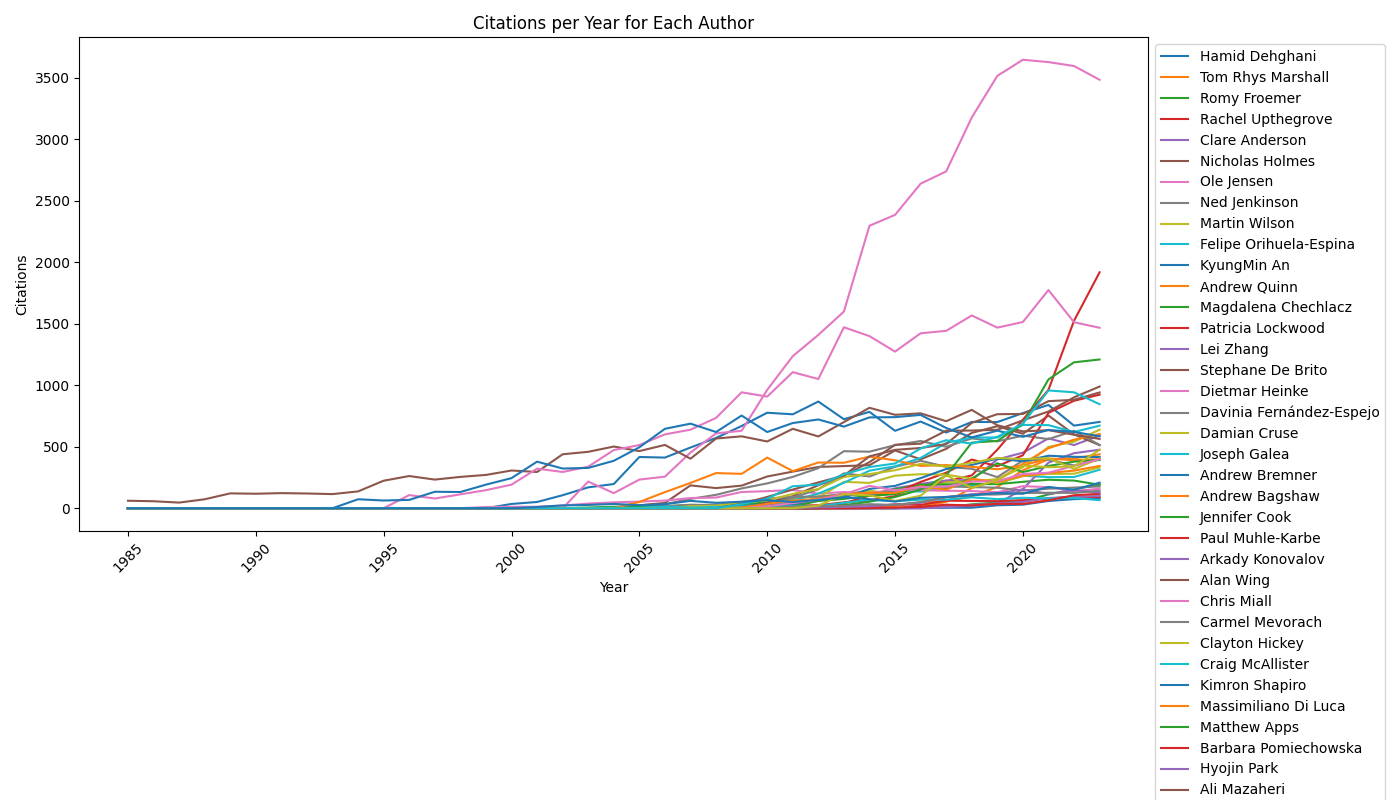
We can then run our predictive model. This function processes the citation data to fit the prophet model, predicts citations for the years 2024, 2025, and 2026, and returns these predictions. After applying the forecasting function to each row in the dataset, the script calculates cumulative citations for each year, including the forecasted years, and appends these cumulative citations to the original dataset.
import pandas as pd
from prophet import Prophet
# Load the data from the CSV file
data = pd.read_csv('../data/citation_stats/citation_statistics.csv')
# Remove the 'citations_2024' column
data = data.drop(columns=['citations_2024'])
# Function to perform forecasting for each person
def forecast_citations(row):
# Extract the relevant citation columns
citation_cols = [col for col in row.index if col.startswith('citations_')]
# Create a dataframe with the citation data
citation_data = pd.DataFrame({'ds': citation_cols, 'y': row[citation_cols]})
# Remove rows with zero citations
citation_data = citation_data[citation_data['y'] != 0]
# Convert the 'ds' column to datetime by extracting the year from the column name
citation_data['ds'] = pd.to_datetime(citation_data['ds'].str.split('_').str[1], format='%Y')
# Create a Prophet model
model = Prophet()
# Fit the model to the citation data
model.fit(citation_data)
# Generate future dates for forecasting
future_dates = pd.DataFrame({'ds': ['2024-01-01', '2025-01-01', '2026-01-01']})
# Make predictions for the future dates
forecast = model.predict(future_dates)
# Extract the forecasted values
predicted_values = forecast['yhat'].values
return pd.Series(predicted_values, index=['prediction_2024', 'prediction_2025', 'prediction_2026'])
# Apply the forecasting function to each row
data[['prediction_2024', 'prediction_2025', 'prediction_2026']] = data.apply(forecast_citations, axis=1)
# Get the citation columns
citation_cols = [col for col in data.columns if col.startswith('citations_')]
# Sort the citation columns in ascending order
citation_cols = sorted(citation_cols, key=lambda x: int(x.split('_')[1]))
# Create a new DataFrame to store the cumulative citations
cumulative_data = pd.DataFrame(index=data.index)
# Calculate the cumulative citations for each year
for i in range(len(citation_cols)):
if i == 0:
cumulative_data[f'cumulative_{citation_cols[i].split("_")[1]}'] = data[citation_cols[i]]
else:
cumulative_data[f'cumulative_{citation_cols[i].split("_")[1]}'] = cumulative_data[f'cumulative_{citation_cols[i-1].split("_")[1]}'] + data[citation_cols[i]]
# Calculate the cumulative citations for the predicted years
cumulative_data['cumulative_2024'] = cumulative_data[f'cumulative_{citation_cols[-1].split("_")[1]}'] + data['prediction_2024']
cumulative_data['cumulative_2025'] = cumulative_data['cumulative_2024'] + data['prediction_2025']
cumulative_data['cumulative_2026'] = cumulative_data['cumulative_2025'] + data['prediction_2026']
# Concatenate the original data with the cumulative citations
updated_data = pd.concat([data, cumulative_data], axis=1)
# Save the updated data to the same CSV file
updated_data.to_csv('citation_statistics_with_predictions.csv', index=False)
After calculating the cumulative citations for the predicted years (2024, 2025 and 2026), we can plot the predicted citation number, as well as the cumulative predicted citations for these years:
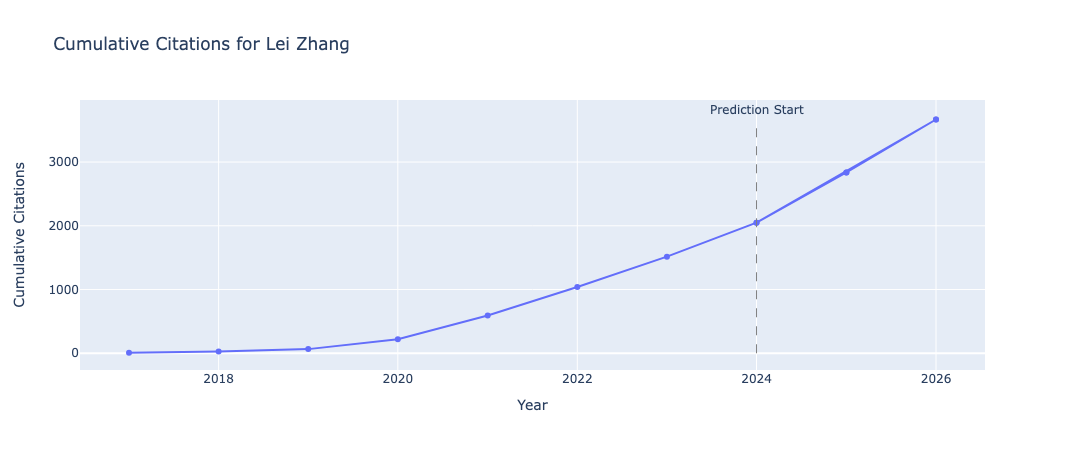
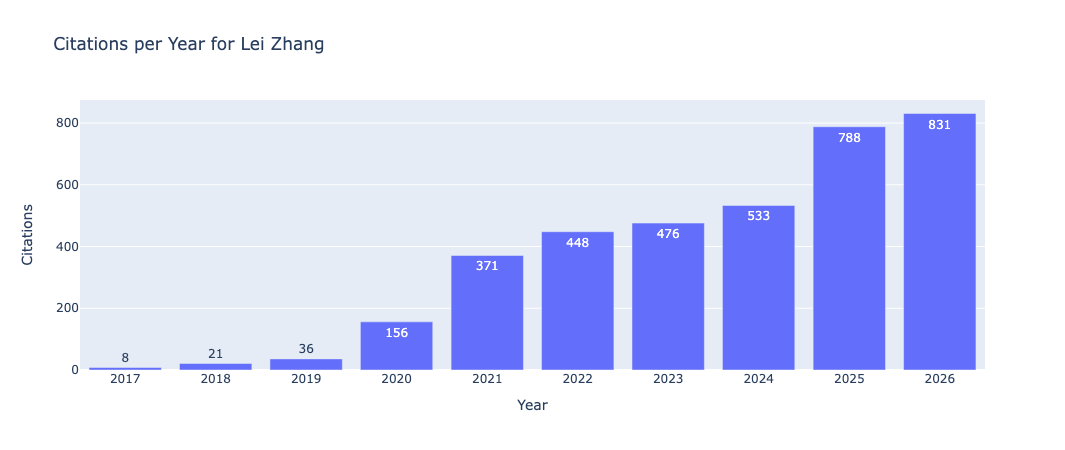
These are displayed using plotly within the Jupyter notebook, which are interactive plots.
How accurate are the predictions?
It’s all well to use a model in making some prediction, but how accurate is that model? The final section of the notebook calculates predictions for the year 2023 (the last year where complete yearly citation data are available) and compares this prediction to the actual values across all researchers.
Ultimately, the model has an accuracy of 80.63%, which whilst good, isn’t great.
Congratulations, you just extracted data from a website, ran forecasting models and created interactive plots!